Definition and Function of Enzymes
Table of Contents
- Definition of Enzymes
- Enzymes as Biocatalysts
- Covalent Modification Control
- The Active Center
- The 4 Enzyme Specificities
- Classification of Enzymes
- Isoenzymes – Same Reaction, Different Structure
- Coenzymes and Prosthetic Groups
- Serine Proteases
- Kinetics of Enzymes
- Regulation of Enzymes
- Enzyme Defects and their Impact
- Review Questions
- References
Definition of Enzymes
Enzymes are biocatalysts, which accelerate chemical reactions and thus play a vital role in human metabolism. Almost always, they consist of proteins with a molecular weight of about 10,000 – 100,000 Daltons and are formed in the cell as part of the protein biosynthesis.
An exception, however, is catalytically active nucleic acids, the so-called ribozymes such as the snRNA. The substances that are converted by enzymes are referred to as substrates, and the end result is the products.
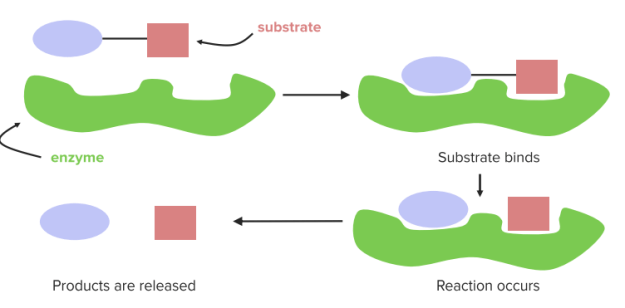
Enzymes as Biocatalysts
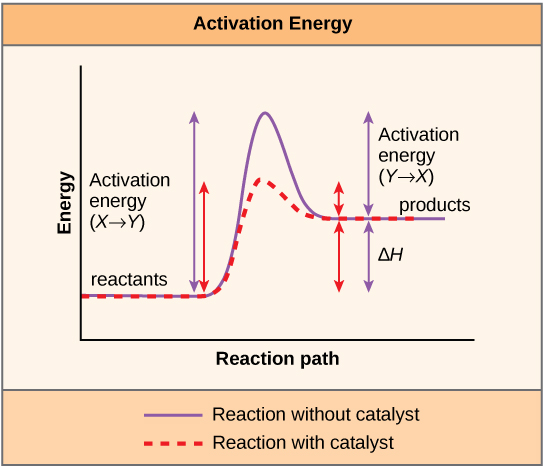
Biocatalysts do not change the reaction pathway of a chemical reaction, but rather they accelerate the reaction by lowering the activation energy. During the conversion of substrates(S) to products (P), the enzyme (E) is not consumed and exists unchanged after the reaction. The reaction pathway can be represented simply as follows:
E + S ↔ ES ↔ E + P
During the reaction of substrates to products, a transition state must be overcome which is unfavorable in comparison to the energetic state of substrate and product, which means, it has a higher free energy. The activation energy is thus the difference between the free energy of substrate and transition state.
In order to reduce the activation energy, enzymes undergo an interaction with substrates, also known as Enzyme-Substrate Complex (ES). This stabilizes the transition state and its free energy is lowered. The substrate binding occurs at the active center of the enzyme.
Covalent Modification Control
Zymogens
Protease precursors
- Pepsinogen
- Proenteropeptidase
- Trypsinogen
- Chymotrypsinogen
- Procarboxypeptidases
- Blood clotting proteins
- Procaspases
- Proelastase
Other
- Pacifastin
- Plasminogen
- Angiotensinogen
- Prolipase
- Pre-proinsulin
Chemical modifications
- Phosphorylation — Kinase cascades
- Acetylation — Histones
- Formylation — All prokaryotic proteins
- Acylation — Anchored membrane proteins (SRC)
- ADP ribosylation — Transcription factors
- Prenylation — Ras
- Sulfation — Serine protease inhibitors
- Ubiquitination — Many proteins
- γ-Carboxylation — Clotting proteins
Gene expression control in hypoxia
- Under conditions of low oxygen (hypoxia), cells produce a transcription factor called HIF-1
- HIF-1 induces expression of genes to deal with the hypoxic conditions
| Transport glucose into cells | |
| GLUT 1 and GLUT 3 | |
| Glycolysis | |
|
|
The Active Center
The active center is a three-dimensional, specifically folded region, made of amino acids and is usually arranged in a hollow shape. The conditions for the chemical reaction are favored within this center, because the probability of the overlapping of the reactants is improved by the convergence and optimal orientation in space, among other things.
The binding of the substrate to the active center is not covalent. The substrate binding is carried out by:
- Ionic interaction
- Hydrophobic interaction
- Van der Waals force
- Hydrogen bridge bonds
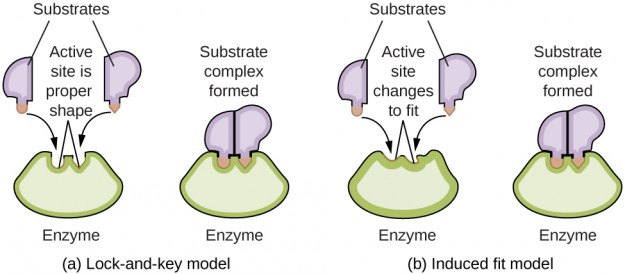
The substrate binding functions according to the induced-fit model, which states that the active center changes its conformity after substrate binding in such a way that it is complementary to the substrate. Previously, it was assumed that the conformity of the active center existed prior to substrate binding, which was referred to as the key-lock principle; this view is, however, outdated.
The 4 Enzyme Specificities
These specificities distinguish the enzymes as biological catalysts from the chemical catalysts (e.g., platinum). The 4 specificities of enzymes are:
1. Substrate Specificity
The induced-fit model forms the basis of the substrate specificity of enzymes and means that each enzyme can convert only one particular substrate; for example, in the context of glycolysis, the glucokinase of the liver converts only glucose into glucose-6-phosphate and no other hexoses.
2. Stereospecificity
Also, the concept of stereoisomerism plays an important role: lactate dehydrogenase, for example, can only convert L-lactate to pyruvate and not its mirror image D-lactate. The lactate dehydrogenase is thus stereospecific, or optically specific.
3. Group Specificity
If an enzyme reacts with a specific chemical group (e.g., amino group) that is located on the substrate, this is called group specificity. Then, the substrate itself is not relevant, but rather the chemical group that it carries.
4. Reaction Specificity
The specificity of reaction states that each enzyme can catalyze only a specific type of reaction. To illustrate: The enzyme X catalyzes the reaction E + S ↔ ES ↔ E + P, but might not catalyze similar reaction E + S ↔ ES ↔ E + P1 + P2.
Classification of Enzymes
Currently, there are more than 2,000 known enzymes. They are divided into 6 main groups, which, in turn, are divided into subclasses. The following table should help to get a good overview:
Isoenzymes – Same Reaction, Different Structure
Some enzymes catalyze the same reaction but nevertheless differ slightly in their structure. Such enzymes are referred to as isoenzymes. Their amino acid sequence is different because they are coded by different genes.
They play an important role in medicine since different isoenzymes are typical for specific organs, and can thus provide conclusions about pathologies. When a certain isoenzyme is increased in the blood, it can be a valuable diagnostic parameter for the detection of a disease.
The best-known example is probably creatine kinase (CK). It consists of 2 subunits, the M-type (muscle) and B-type (brain), and has 3 isoforms. An increase of the CK-MB isoform, for example, can be traced back to a heart attack since this enzyme is mainly localized in the heart muscle.
Another example would be lactate dehydrogenase (LDH). It consists of four subunits, which are composed of the H-type (heart) and M-type (muscle). In the myocardium, one finds the isoenzyme LDH1 which consists of 4 H subunits, whereas in the skeletal muscle, one finds the isoenzyme LDH5 with 4 M subunits.
Coenzymes and Prosthetic Groups
However, enzymes are often dependent on auxiliary molecules, so-called coenzyme or cosubstrates, without which a reaction might not be catalyzed. These helpers can…
- …be firmly bound to the enzyme and are then called a prosthetic group.
- …exist in a soluble form.
Their purpose is to support the enzyme by taking over, for example, the transfer of electrons. The following table provides an overview of the most important coenzymes:
Some enzymes, such as α-ketoglutarate dehydrogenase of the citric acid cycle, require several coenzymes; in this case, for example, next to CoA also thiamine pyrophosphate, lipoamide, FAD, and NAD+.
Serine Proteases
- Cleave peptide bonds
- Specificity of cutting
- Common active site composition/ structure
- Mechanistically well studied
Kinetics of Enzymes
Enzyme kinetics describes the sequence of enzyme-catalyzed reactions with the dependence on various parameters.
Dependence of the reaction rate on temperature
One should remember the following thumb rule here: The reaction rate doubles for every 10-degree increase in temperature. This can certainly only take place within a limited scope since enzymes have an optimum temperature and denature at too high temperatures.
Dependence of the reaction rate on pH value
There is no thumb rule as the one for temperature dependence. However, it is important to know that enzymes have a pH optimum and changes in the pH value cause functional groups of the enzyme itself, or of its substrate to change.
This leads to changes in the spatial structure so that the bond at the active center is improved or deteriorated. In the case of stronger pH changes, it may even come to denaturation of the enzyme. The enzymes of the stomach represent a peculiarity in this regard: Pepsin, for example, works effectively at a pH of about 2, where other enzymes would long have denatured.
Dependence of the reaction rate on substrate concentration
1. Michaelis-Menten model
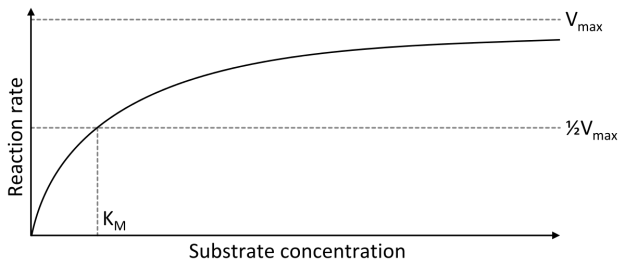
Image: “Michaelis-Menten-Diagramm” by Thomas Shafee. License: CC BY-SA 4.0
The Michaelis-Menten model has become the established explanation for this relation. If one plots the reaction rate of the reaction E + S ↔ ES ↔ E + P against the substrate concentration on a graph, the result is a hyperbolic curve, which approaches the maximum velocity vmax.
A simple example helps to understand the graph: If you have 5 enzymes and add 0 substrates, the reaction rate is zero, since there is nothing to be converted. If you now gradually add more and more substrate, the reaction rate approaches the maximum velocity. If all 5 enzymes are occupied with substrates, one speaks of substrate saturation and the maximum velocity is reached.
An important concept in this context is the Michaelis-Menten constant (KM), as it corresponds to the substrate concentration at half-maximal velocity (½ vmax). This means that, at this time, half of all enzymes are occupied with the substrate, i.e. they form part of an enzyme-substrate complex. The Michaelis-Menten constant thus indicates the affinity of an enzyme for its substrate and is characteristic for the particular enzyme-substrate complex.
Explanation: If the KM value is low, this means that the half-maximal velocity is present, even when the substrate concentration is low and that the enzyme is thus strongly affine for this substrate. If the KM value is large, however, then the half-maximal velocity will only be achieved if the range of substrates is greater, since the enzyme is less affine for the substrate. It follows the smaller the KM is, the stronger the reaction rate increases with the substrate concentration.
The Michaelis-Menten constant depends on the temperature and pH value, but is independent of the enzyme concentration.
For calculating the reaction rate of enzyme-catalyzed reactions in dependence of the substrate concentration, the Michaelis-Menten equation is used. It states:
Now there are 3 possibilities:
- The substrate concentration S is significantly lower than KM. In this case, one can neglect the addition of S to KM, as this would not significantly increase the value of the denominator and one obtains the equation: v = vmax [S]/KM
- The substrate concentration is equal to KM. In this case, one can summarize the denominator KM+ [S] to 2 [S], so that the equation now reads v = vmax · [S]/2[S] and this can be shortened to v = ½ vmax
- The substrate concentration is significantly higher than KM. In this case, one can neglect KM during the addition in the denominator, so that the equation is v = vmax · [S]/[S] and the fraction can be shortened to v = vmax. This is due to substrate saturation when there are very high substrate concentrations, and therefore maximum velocity is achieved.
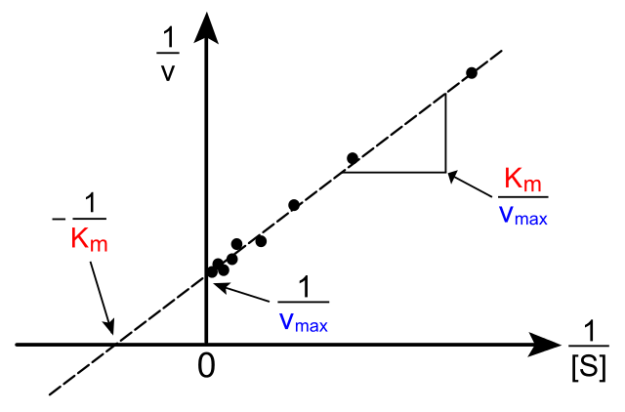
2. Lineweaver-Burk graph
Also, the following graph is important to know in exams. It is merely a different representation of the reaction rate, depending on substrate concentration. Through the complicated transformation of the Michaelis-Menten equation, a typical linear equation of the form y = mx + n is derived. It states:
Regulation of Enzymes
The human body has the ability to stimulate or to curb metabolic processes through the regulation of enzymes. This regulation can be long- or short-term in nature.
Short-term regulatory mechanisms are:
- Competitive inhibition
- Non-competitive inhibition
- Uncompetitive inhibition
- Interconversion
- Allosteric regulation
The long-term regulatory mechanisms, in turn, include:
- Increased synthesis of necessary enzymes
- Increased reduction of unnecessary enzymes
Inhibition of Enzymes
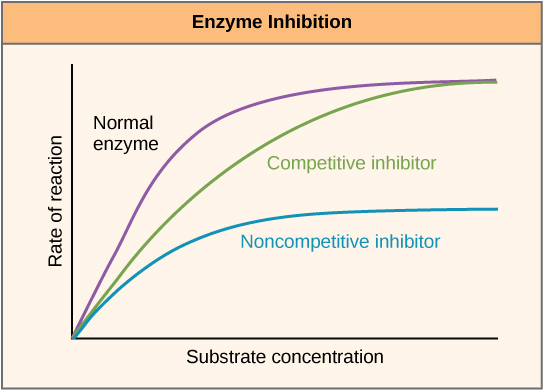
One distinguishes 3 types of enzyme inhibition:
In competitive inhibition, the inhibitor resembles the substrate and thus competes with it for the binding site at the active center of the enzyme. The inhibitor binds at the active center, thereby blocking this binding site. The KM value increases since an increased substrate concentration is required in order to obtain the half-maximal velocity.
Vmax is however not changed. The inhibitor can be removed from the bond through an excess of the substrate; therefore, this inhibition is reversible. Examples: ACE inhibitors, Curare.
In non-competitive inhibition, the inhibitor does not have to have a similar substrate because it binds and inhibits to the enzyme outside of the active center. The substrate can thus continue to bind to the active center but is not converted due to the additional binding of the inhibitor. This means that the number of functional enzyme-substrate complexes decreases.
This decreases Vmax while KM remains the same. A substrate excess does not displace the inhibitor. The non-competitive inhibition can be reversible or irreversible. Example: acetylsalicylic acid (irreversible).
Uncompetitive inhibition is a rare form of enzyme inhibition and characterized by specific binding at the enzyme-substrate complex. The inhibitor also binds outside the active center, but only if the enzyme-substrate complex is already formed. The result is a reversible conformational change, and thus inactivation of the enzyme. KM and vmax are reduced.
Irreversible Enzyme Inhibition: Suicide – Enzyme Inhibitors
Allosteric Effects
Allosteric regulation is the most common form of regulation, and takes place with the help of allosteric ligand/effectors. These bind outside the active center of the enzyme and namely at the allosteric center. This leads to a conformational change and thus, activation or deactivation of the enzyme.
In allosteric enzymes, 2 state forms can be distinguished: the inactive T-form (tensed) and the active R-form (relaxed). Allosteric inhibitors thus stabilize the T-form and allosteric activators stabilize the R-form. Often, the substrate itself represents an allosteric activator, which makes sense because the substrate thereby promotes the R-condition for its own implementation.
Depending on the nature of allosteric effector, the maximum velocity, or the KM value, is changed. Effectors of the v-type increase/decrease the maximum velocity, and effectors from the k-type influence the KM value. However, the dependence of the reaction rate on substrate concentration in allosteric enzymes is not hyperbolic, but rather sigmoid.
This is due to the fact that allosteric enzymes usually consist of several subunits and hence have more active centers, so that the binding of the first substrate, which at the same time is also the allosteric activator, facilitates the binding of the second substrate. This effect is called positive cooperativity.
However, if the binding of the first substrate complicates the binding of the second substrate, this is called negative cooperativity. Cooperativity can not only be found in enzymes, but also in, e.g., hemoglobin.
Interconversion
Enzymes can also be regulated by interconversion. This means that specific groups (e.g., a phosphate residue) are added to, or cleaved off, of the enzyme, which changes the active state of the enzyme. The phosphorylation of the enzyme is mediated by kinases, and dephosphorylation by phosphatases.
Enzyme Defects and their Impact
Phenylketonuria
Enzymes can be limited in their activity, or fail completely, due to, e.g., a genetic defect. One example of such an enzyme deficiency disease is phenylketonuria (PKU). In this disease, the enzyme phenylalanine hydroxylase is defective, so that phenylalanine cannot be converted into tyrosine.
The accumulation of phenylalanine leads to the formation of neurotoxic substances, which, if left untreated, results in mental retardation with increased convulsions beginning in the 4th month of life. Treatment of choice is a controlled diet with reduced phenylalanine contents. If this is already started in infancy, the mental retardation can be prevented completely.
Myasthenia Gravis
Image: “Patient with Myasthenia Gravis” by Phil Schatz. License: Public Domain
Sometimes, it can be necessary to actually restrict the enzymatic activity. This would be the case with the myasthenia gravis disease, which manifests as increased muscle fatigue.
The disease is caused by antibodies that are directed against the acetylcholine receptors of the muscle endplate and block them. Treatment, therefore, consists in the administration of acetylcholinesterase inhibitors such as pyridostigmine, so that the degradation of the transmitter acetylcholine in the synaptic cleft is reduced, maintaining higher concentrations at the muscle endplate in order to trigger endplate potentials.
Review Questions
The answers can be found below the references.
1. Which statement is incorrect? The substrate binding is carried out via…
- …Van-der-Waals forces.
- …atomic bonding.
- …hydrogen bonding.
- …hydrophobic interaction.
- …ionic interaction.
2. Which statement is false?
- Oxidoreductases transfer reduction equivalents.
- Hydrolases separate molecules under hydration.
- Ligases form energy independent bonds.
- Aminotransferases transfer amino groups.
- Lyases split energy independent bonds by introducing double bonds.
3. Which statement is correct for allosteric regulation?
- Allosteric effectors always have an activating effect.
- Allosteric enzymes are active in the T-shape.
- Allosteric effectors never affect the KM value.
- Allosteric regulation occurs only rarely.
- The substrates themselves are often allosteric ligands.





Comentários
Enviar um comentário