Single Nucleotide Polymorphisms and BRCA 2 Gene
Table of Contents
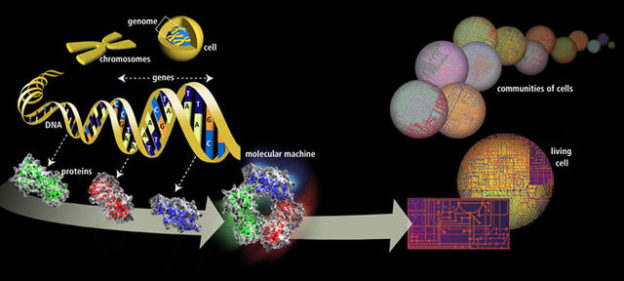
Image: “From DNA to Life” by Dodo. License: Public Domain
Introduction to Genomes
The complete set of the genetic material of an organism is called a genome. A genome consists of DNA (or RNA for RNA viruses) which includes the genes (coding region), non-coding DNA, and DNA (or RNA) of mitochondria as well.
The term genome was coined in 1920 by Hans Winkler. This term is a combination of genes and chromosomes.
Inside the nucleus of a human cell are 23 pairs of chromosomes containing DNA strands. Each arm DNA consists of four nucleotide bases (adenine (A), thymine (T), guanine (G) and cytosine (C)) which are arranged in a specific sequence that determines the genes.
A genome contains all the necessary information that is needed by an organism to produce, maintain, and reproduce. Each genome in the human body contains more than 3 million of DNA base pairs, and all of this fits inside the microscopic nucleus of every cell.
Number of Genes and Complexity
The number of genes does not determine the complexity of an organism. There are 50,000 genes in corn, 45,000 in rice, up to 25,000 genes in humans and 13,600 genes in a housefly.
Gene Location
Genes can be located on a DNA strand by searching for start codon, stop codon, and open reading frame (ORF) which is the region between the start and stop codon.
Sequence inspection
Genes are not a series of random nucleotide sequences, rather, they have a specific sequence and feature; thus, sequence inspection can be done for locating genes on DNA. The particular sequence and features help determine whether a given sequence is a gene or not.
Sequence inspection is usually the first method for analyzing gene sequence. It is not a foolproof method of analysis but, undoubtedly, a useful tool for locating genes.
Open Reading Frames ORF
ORF begins with a start codon, which has the sequence of nucleotide base as ATG and finishes at end codon consisting of a TAG, TAA or TGA. Each strand of DNA has three reading frames in one direction and three on the other strand in the opposite direction; thus, both the strands will have six reading frames.
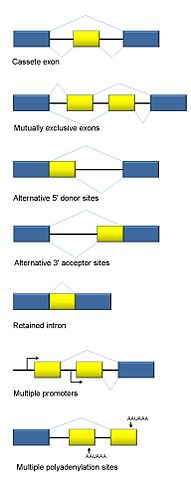
Alternative Splicing
Image: “Collection of basic types of alternative RNA splicing events” by Agathman. License: CC BY-SA 3.0
The information stored in the genes on DNA is transcribed into mRNA and then translated into proteins. However, before the formation of proteins, the introns (non-coding parts of a gene sequence) must be removed, and the exons (coding parts of proteins) of mRNA must be joined for it to translate into proteins.
Alternative splicing is a regulated method during gene expression that results in the formation of multiple proteins from a single gene. This involves inclusion or exclusion of a particular exon from the final mRNA, and results in the formation of a variety of proteins with different amino acid sequence and different functions. Thus, even if the number of genes in an organism is less, the room for complexity may largely be increased due to alternative splicing of mRNA.
Modes of Alternative Splicing
Five modes of alternative splicing are:
- Exon Skipping (Cassette Exon)
- Mutually exclusive exons
- Alternative donor site
- Alternative acceptor site
- Intron retention
Types of DNA Sequences
The method of determining the exact order of nucleotide bases in a DNA is called DNA sequencing.
The types of DNA sequences include:
Coding DNA sequences
- Single copy genes
- Segmental duplications
- Multigene families
- Tandem clusters
Non-Coding DNA sequences
- Intron
- Structural DNA
- Simple sequence repeats
- Segmental duplications
- Transposable elements
- Pseudogenes
- Micro RNAs
- Long non-coding RNA
Coding DNA Sequences
These are the regions of DNA that code for proteins.
Single copy genes
They are transcribed to form RNA which is translated to form proteins.
Segmental duplications
They are long DNA sequences and are almost identical (90% – 100%) in sequence. They are present in multiple locations because of duplicating events. Can be tandem or interspersed and interchromosomal or intrachromosomal.
Multigene families
It includes a group of genes from the same organism that forms proteins with a similar order either over the full length of the gene sequence or over a partial domain. DNA duplication can form gene pairs, and a multigene family will form if both copies in subsequent generations exist. Genes that encode for hemoglobin, actins, interferon, histones, etc. are examples of multigene families.
Tandem genes and cluster genes
Tandem genes are present within the segment of DNA that are repeated a number of times from head to tail. Cluster genes are connected by non-conserved DNA, but irregularly spaced and inverted unpredictably.
Non-Coding DNA Sequences
These are the regions of DNA that do not code for proteins.
Introns
It is a nucleotide sequence of a gene in DNA or RNA that is not included and spliced out during final formation of mRNA and does not code for protein formation.
Pseudogenes
They are genes that do not code for proteins due to mutations like frameshift or premature stop codons.
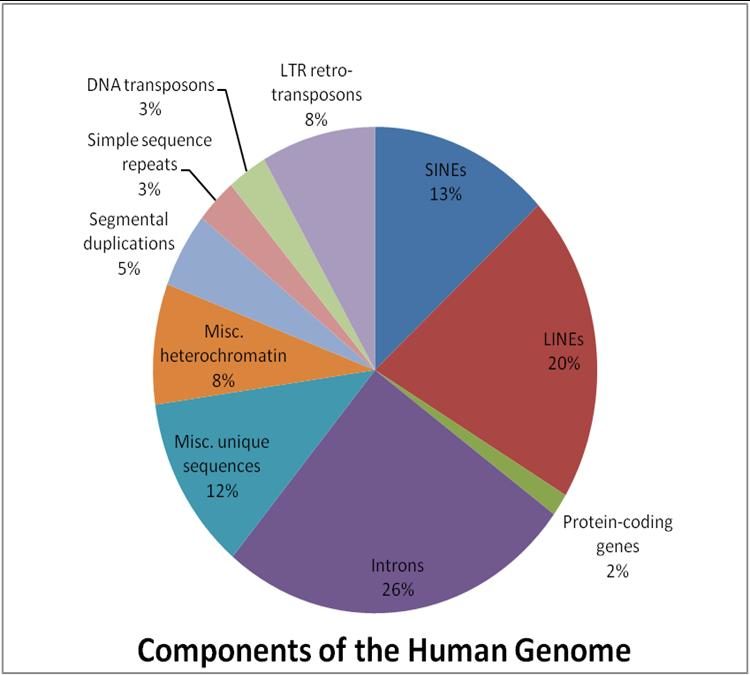
Transposable elements
Also called transposons or jumping genes, they are the sequence of genes on DNA that move from one place to another within a genome. They can be duplicated or excised and inserted elsewhere. They include Long Interspersed Elements LINEs (21%), Short Interspersed Elements SINEs (13%), Long Terminal repeats LTRs (8%) and Dead Transposons (3%).
Transposable elements
These elements move from one location to another; they may be:
- Duplicate
- Excised
If excised, these elements are inserted somewhere else.
Types of transposable elements
1. Dead transposons: This 3% of the genome has no machinery to move.
2. Long terminal repeats LTRs: 8% of human genome is LTRs. They contain reverse transcriptase.
3. Short interspersed elements (SINEs): 13% of the human genome consists of SINEs. These are nested in long interspersed elements.
4. Long interspersed elements (LINEs): They make up to 21% of the human genome. LINEs can transpose themselves.
The remaining 55% makes the non-coding and coding DNA in the human genome.
Image: “This figure illustrates the proportion of various genomic components within the human genome.” by Alglascock. License: CC BY-SA 3.0
Single Nucleotide Polymorphism
Single nucleotide polymorphism (SNP) is the most common type of DNA sequence variationthat occurs when a single nucleotide (A, T, G, or C) varies between members of a species or even between paired chromosomes within a person. These changes may be responsible for diversity among people and some common familial traits like diabetes, hypertension, curly hair, drug response, etc.
SNPs are used to tag genotypes. Known SNPs have been mapped onto genome sequences.
SNPs are found in a human DNA. They occur in every 300 nucleotides. They play an important role as biological markers to locate genes in diseased condition and by directly affecting the gene’s function. According to recent research works, SNPs may contribute in judging the individual response to certain drugs, and certain environmental factors such as toxins and risk of developing disease. It can tract the inheritance of disease genes in families.
BRCA2 Gene
This gene instructs the formation of tumor suppressor protein which helps prevent cells from uncontrollable growth and multiplication. This protein is also involved in repairing the damage caused by factors like radiation or some environmental exposures; thus, this protein (and, hence, the gene) aims at maintaining and preserving the genetic information.
Expressed Sequence Tag (EST)
The EST is a part of cDNA sequence in the form of a short sub-sequence. In gene-sequence determination and gene discovery, Expressed Sequence Tag is instrumental. These are also used for the identification of gene transcripts.
In genetics, EST helps determine which pieces of the genome are expressed.
Microarray
A microarray contains catalogued genes from an entire genome. They can tell when a gene is expressed.
Comentários
Enviar um comentário